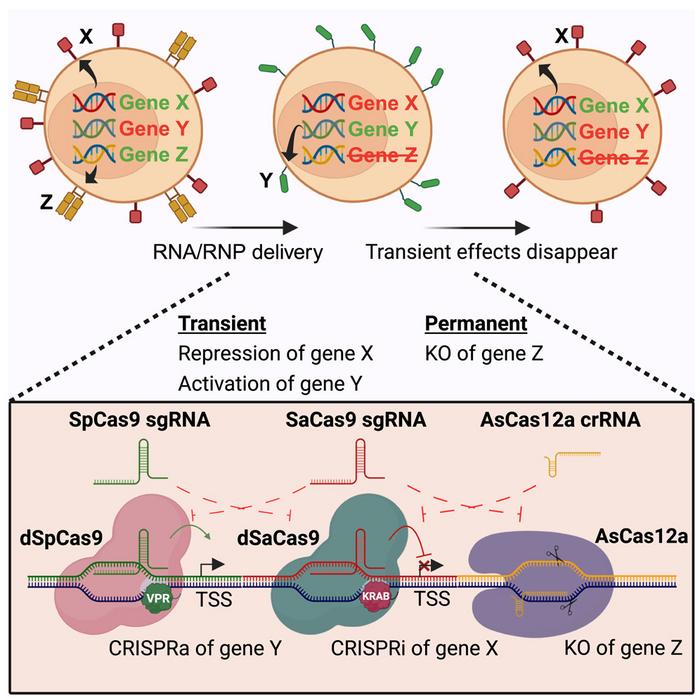
Researchers at Aarhus University in Denmark have developed a new method to achieve precise and complex genetic manipulations by combining different CRISPR/Cas systems, each specialised for a different function. They used nuclease-deficient Cas9 (dCas9) from Streptococcus pyogenes fused with the VPR activation domain, dSpCas9-VPR, for temporary gene activation, dCas9 from Staphylococcus aureus fused with the KOX1 repressor domain, dSaCas9-KOX1, for temporary gene repression, and Cas12a from Acidaminococcus (AsCas12a) or SpCas9 for permanent gene deletion.
Using orthogonal CRISPR/Cas components, the team demonstrated that the trimodal genetic engineering system can simultaneously activate, suppress, and permanently delete different genes within the same cell. According to the authors, this approach has immense potential for cell therapy manufacturing and regenerative medicine by enabling complex genetic engineering and precise control over cell behaviour and function.
»This one-step approach combines transient transcriptome modulation with permanent genomic modifications, holding significant promise for applications like cell therapy and regenerative medicine, where synergistic effects of transient and permanent modifications can improve therapeutic outcomes,« said Amalie Dyrelund Broksø, co-first author of the study, which was recently published in Molecular Therapy.
Need for complex genetic engineering methods
Although CRISPR/Cas9 has opened new avenues for the treatment of genetic diseases through genetic engineering, most therapeutic applications of CRISPR focus on modifying one gene at a time. However, many biological processes and diseases involve multiple genes working in concert, and the ability to simultaneously control numerous genes in different ways, such as temporarily activating or silencing some genes while permanently deleting others, is challenging. This unmet need is critical for developing sophisticated therapeutic strategies, particularly for diseases involving complex genetic interactions. The research team, led by Dr. Rasmus O. Bak at Aarhus University, aimed to address this gap by exploring the use of orthogonal CRISPR/Cas systems to achieve trimodal genetic engineering.
»Our motivation was to explore the combination of multiple CRISPR/Cas systems to enhance the versatility and precision of genetic engineering,« Broksø explained. »We optimised CRISPR activation and interference for simultaneous use, enabling transient gene regulation in single cells. Adding CRISPR-based gene editing introduced a new layer of functionality, resulting in a ‘trimodal engineering’ framework.«
Screening of CRISPRa and CRISPRi effectors
The team first optimised each component separately by testing different versions in Jurkat cells to identify the most effective effectors for CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi).
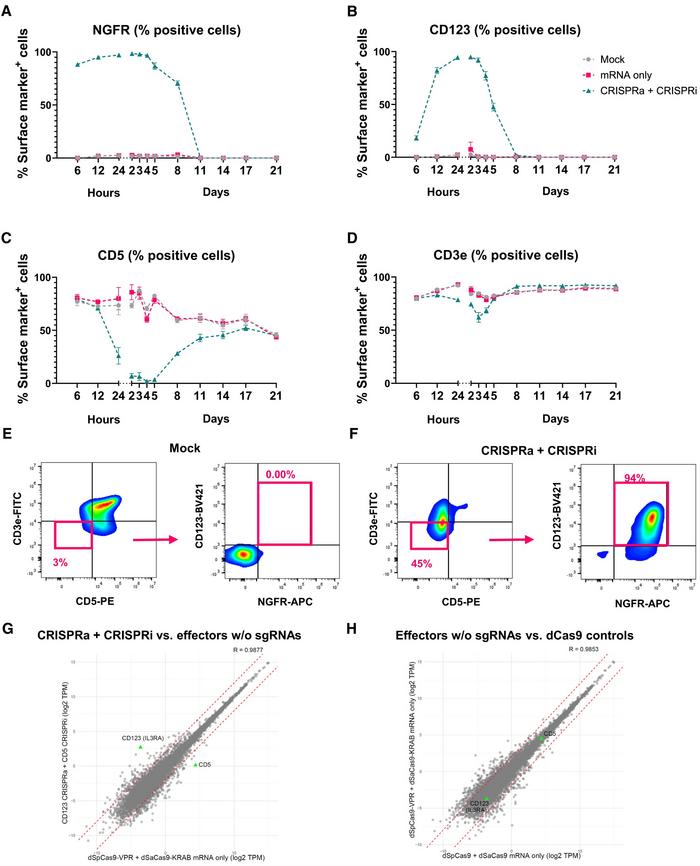
For CRISPRa, they tested seven different constructs, including direct fusions to dSpCas9 (VPR, VPRmini, and sp65p3-HSF1) and SunTag-based systems (scFv-VP64, scFv-VPR, scFv-VPRmini, and scFv-sp65p3-HSF1). The efficiency of gene activation was assessed for multiple target genes (NGFR, CD123, CD19) using flow cytometry. These genes were selected as they encode surface proteins that are easily detectable by flow cytometry. The dSpCas9-VPR fusion consistently showed the highest efficiency for gene activation across all target genes tested. However, transcription activation varied depending on the target gene (ranging from ~75% of positive cells for CD19 to 90%–95% for NGFR and CD123), highlighting the importance of optimising CRISPRa systems for specific applications.
»The difference in optimal conditions does imply that the system is not as ‘plug-and-play’ as it might seem,« explained Louise Bendixen, co-first author of the study. »It could potentially need to be optimised for the specific mix of target genes, effectors, and cell types used in the therapy during development.«
CRISPRi effectors were evaluated using both dSpCas9 and dSaCas9 fused to various repressor domains (KOX1, ZIM3, and KOX1-MeCP2). The efficiency of gene repression was tested on CD5 and CD3e — which encode proteins regulating the immune system — in Jurkat cells. With dSpCas9, all three repressors showed similar potency in suppressing CD5 and CD3e expression (~10% of CD5-positive cells and ~5% for CD3e-positive cells). However, with dSaCas9, KOX1 demonstrated superior repression, particularly for CD3e (CD5: ~5% of CD5-positive cells with KOX1 vs 5%–10% with ZIM3 and KOX1-MeCP2; CD3e:
Multiplexed transcriptional modulation
The most effective CRISPRa and CRISPRi systems were combined to achieve simultaneous activation and repression of different genes. The performance of the system was tested by activating NGFR and CD123 and repressing CD5 and CD3e in Jurkat cells.
Using the identified optimal effectors (dSpCas9-VPR for activation and dSaCas9-KOX1 for repression), the team achieved simultaneous activation of NGFR and CD123 and repression of CD5 and CD3e in Jurkat cells. The effects were transient but robust, with NGFR activation evident within 6 hours post-delivery, peaking between days 1 and 3, with up to 99% of the cells expressing NGFR (Figure 1). CD123 activation showed a slower onset but reached up to 98% positive cells. CD5 repression was nearly complete from days 2–5, while CD3e showed moderate repression. Transcriptional modifications were transient and returned to baseline within 8–21 days.
Efficiency of trimodal genetic engineering
Along with CRISPRa and CRISPRi, the researchers of the present study have integrated gene knockout capabilities into the system using either AsCas12a or SpCas9. This approach allows simultaneous gene activation, repression, and knockout in the same cells.
»Utilising different Cas orthologues for multiple functionalities allows us to perform targeted up- and down-regulation with the assurance that the Cas proteins will only form complexes with the sgRNAs intended for that specific Cas orthologue,« Bendixen explained. »Because there is no cross complexation across Cas orthologues, we can deliver all reagents for three separate CRISPR functionalities together, thus limiting delivery to one electroporation and limiting the impact on cell viability.«
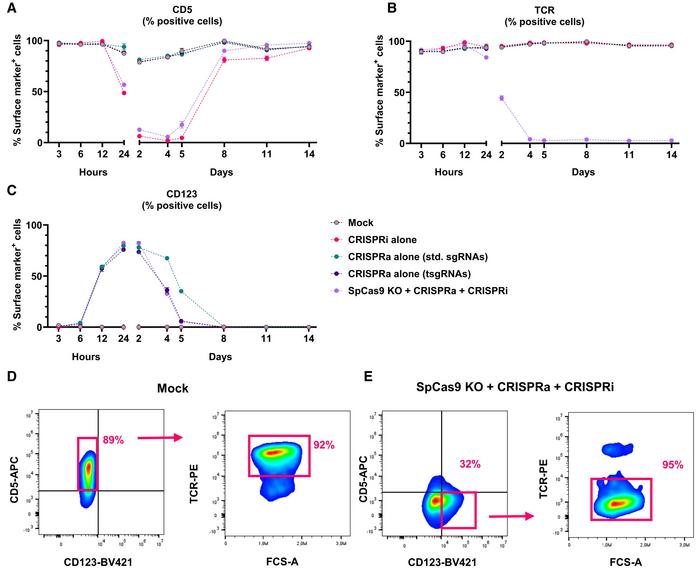
Initially, the team used three different Cas orthologues: dSpCas9-VPR for CD123 activation, dSaCas9-KOX1 for CD5 repression, and AsCas12a for TRAC (T cell receptor alpha chain) knockout. This approach achieved efficient trimodal engineering, with approximately 65% of cells showing the desired expression pattern (CD5-/CD123+/TCR-), compared to 0.01% in the control cells.
To simplify the system, the team developed a two-orthologue approach using truncated guide RNAs (tsgRNAs) with SpCas9, allowing them to use the same type of Cas9 protein for both activation and deletion without interference between the two functions. Although tsgRNAs showed slightly reduced CRISPRa efficiency compared to standard guides (~90% of CD123+ cells with sgRNAs vs 85% % of CD123+ cells with tsgRNAs), they successfully prevented DNA cleavage while maintaining transcriptional activation. They found that the ratio of guide RNA to Cas9 protein was crucial for optimal performance, with a 1.2:1 or 1.5:1 ratio providing the best balance between knockout and activation efficiencies.
»We observed that the truncated guides caused a shorter duration of activation in relation to their full-length counterparts while having little to no effect on activation efficiency,« Bendixen said. “We suspect the truncation reduces the interaction between dCas9 and the target DNA in the cell, leading to a shorter window of activation, which could be utilised for targeting genes that you only want to activate for a shorter period of time.«
The optimised trimodal system was tested in primary human T cells to assess its efficiency and effect on cell viability and function. The team delivered the components into cells via electroporation using mRNA and ribonucleoprotein complexes, avoiding DNA-based delivery methods that could cause unwanted permanent changes. They targeted genes relevant to T cell function, namely CD123 for activation, CD5 for repression, and TRAC for deletion.
»RNA delivery of the CRISPR/Cas components of this system is ideal as we want the transient effect of the CRISPRa and CRISPRi effectors,« Bendixen noted. »The RNA is quickly translated and less toxic to the cells than plasmid, which also carries the risk of integration into the cell genome.«
In primary T cells, up to 98% of cells showed activation of CD123 transcription during days 1–3, approximately 89% of cells demonstrated repression of CD5 by day 2, and 92% of cells showed successful deletion of the TRAC gene by day 4 (Figure 2). Approximately 44% of primary T cells harboured all three genetic modifications (CD5-/CD123+/TCR-) simultaneously, 3 days after electroporation. The effects of gene activation and repression were temporary, lasting 5–10 days, whereas gene deletion was permanent.
Safety and off-target effects of trimodal genetic engineering
The researchers evaluated the transcriptome-wide effects of the orthogonal CRISPRa/CRISPRi system using RNA sequencing to assess the specificity of trimodal genetic engineering and to identify potential off-target effects. These analyses revealed that the system had high specificity, with minimal unintended effects on other genes. RNA sequencing analysis showed that the genetic modifications were precise and did not significantly affect global gene expression in the genetically modified cells. The few differentially expressed genes identified were likely due to the downstream effects of target gene modulation rather than direct off-target editing.
The trimodal genetic engineering approach did not significantly affect T cell viability, proliferation, or CD4/CD8 T cell ratio. Although cytokine production was largely preserved, a modest reduction (1.7-fold) in TNF-α secretion was observed in engineered cells upon stimulation.
Challenges and future directions
This study represents a significant advancement in genetic engineering capabilities, demonstrating that precise, multimodal gene control is possible within single cells. However, though the peak efficiency of each mode was high, the efficiency of achieving all three modifications simultaneously (44%) could be improved, and further optimisation may be required to reduce variations in the timing and duration of temporary modifications between different genes. In addition, the system needs to be tested in more cell types and with different gene combinations.
»Given that this study shows a proof-of-concept for simultaneous orthogonal CRISPR/Cas gene engineering, a large hurdle would be the implementation and optimisation of the system into the specific cell type,« Bendixen noted. »As with general CRISPR/Cas gene editing, the sgRNA design, validation, and off-target analysis would also need extensive optimisation to make the system effective and safe in a therapeutic setting.«
The researchers acknowledge additional challenges that need to be addressed. »One of the challenges we encountered while implementing the trimodal genetic engineering approach was observing differences in activity between SaCas9 and SpCas9«, Broksø explained. »These variations highlighted the importance of selecting the appropriate Cas ortholog for specific applications. Additionally, the differences in characteristics, such as kinetics and the amplitude of gene regulation, suggest that further investigation into other Cas orthologs could enhance our understanding of their efficiency and potential advantages.«
Looking ahead, the researchers see exciting possibilities for therapeutic applications. »One example is the potential application in CAR T-cell therapy,« said Broksø. »The application of gene editing could provide cells with permanent expression of CAR and loss of inhibitory receptors while transient transcriptional manipulations might provide resistance to T cell differentiation and exhaustion related to the manufacturing process or even reprogram cells into more potent T memory stem cells subsets.«
As Bendixen concluded, while implementation of this system would need optimisation of design and conditions, »the system could very well have the potential to improve cell viability, differentiation, and longevity while performing CRISPR/Cas-based gene therapy.«
Link to the original article in Molecular Therapy:
Orthogonal transcriptional modulation and gene editing using multiple CRISPR-Cas systems
Funding statement: The work described herein was funded by a Lundbeck Foundation Fellowship (R238-2016-3349) and by the EU Commission in the form of an ERC starting grant (HSC-CRISPR, project 101041231, Horizon Europe Pillar I). Views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Health and Digital Executive Agency (HADEA). Neither the European Union nor the granting authority can be held responsible for them.
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.

